Distance transform colocalization HOWTO¶
Loading data¶
From the terminal (cmd-space terminal on OSX)
run:
pymeimage path/to/filename.tif
Alternatively, just run pymeimage without arguments and use the file open
dialog which is displayed to find and open your image. pymeimage should be
able to read anything that bioformats can, and a couple of extras.
The viewer¶
You should get an image viewer like the one shown below. There is currently support for 3 dimensions + colour, and the 3rd dimension can be either time or Z (i.e. XYZC or XYTC). The red lines in the histograms on the right adjust the display scaling. Clicking on one of these histograms and pressing ‘m’ will scale that channel so that the display stretches between the minimum and maximum value. Pressing ‘p’ will map the scaling to the 1st and 99th percentile of the data. Pressing the ‘stretch’ button will map all channels min-max. The dropdowns underneath the ‘stretch’ button select whether you are viewing an XY, XZ, or YZ slice, and the scaling of the image. Dragging the ‘Pos’ slider at the bottom of the image will set the current slice in the stack.
Loading the colocalization module¶
pymeimage doesn’t load all it’s modules at startup. To load the
colocalization module, choose coloc from the Modules menu.
Choosing thresholds¶
The distance transform module uses a threshold on the reference channel to determine what belongs to that channel and what doesn’t. Enable threshold mode by checking the box towards the bottom of the display settings panel. That should change the display to look like this.
The double red lines get replaced with a single line which now represents the threshold, and the display shows the thresholded masks. The thresholds can either be set manually by dragging the lines, or by using one of 2 automatic threshold algorithms, represented by the Isodata and Signal fraction buttons. Isodata uses the standard isodata algorithm, whereas signal fraction calculates the threshold that would be needed to capture a given (default 80%) percentage of the signal.
Performing the analysis¶
Once you are happy with the thresholds, choose Processing->EDT Colocalisation from the menu. NB: this won’t show up unless you have loaded the coloc module. You should get the dialog shown in the next figure. You can choose which channel to use as a reference (1:sup:st channel) and which to measure (2:sup:nd channel), as well as the bin sizes for the histogram. If you have more than 2 channels, make sure you pick the right ones.
When you click OK, it will calculate a distance transform from the mask of the first channel and measure the distribution of the (unthresholded) second channel with respect to that mask. It then swaps the order, and calculates the distribution of the first channel with respect to the second channel’s mask. Note that if you have a stack (either 3D or time series) ``pymeimage`` will assume it is a z-stack, and calculate distances in 3D.
When calculations are complete, 3 windows will be displayed.
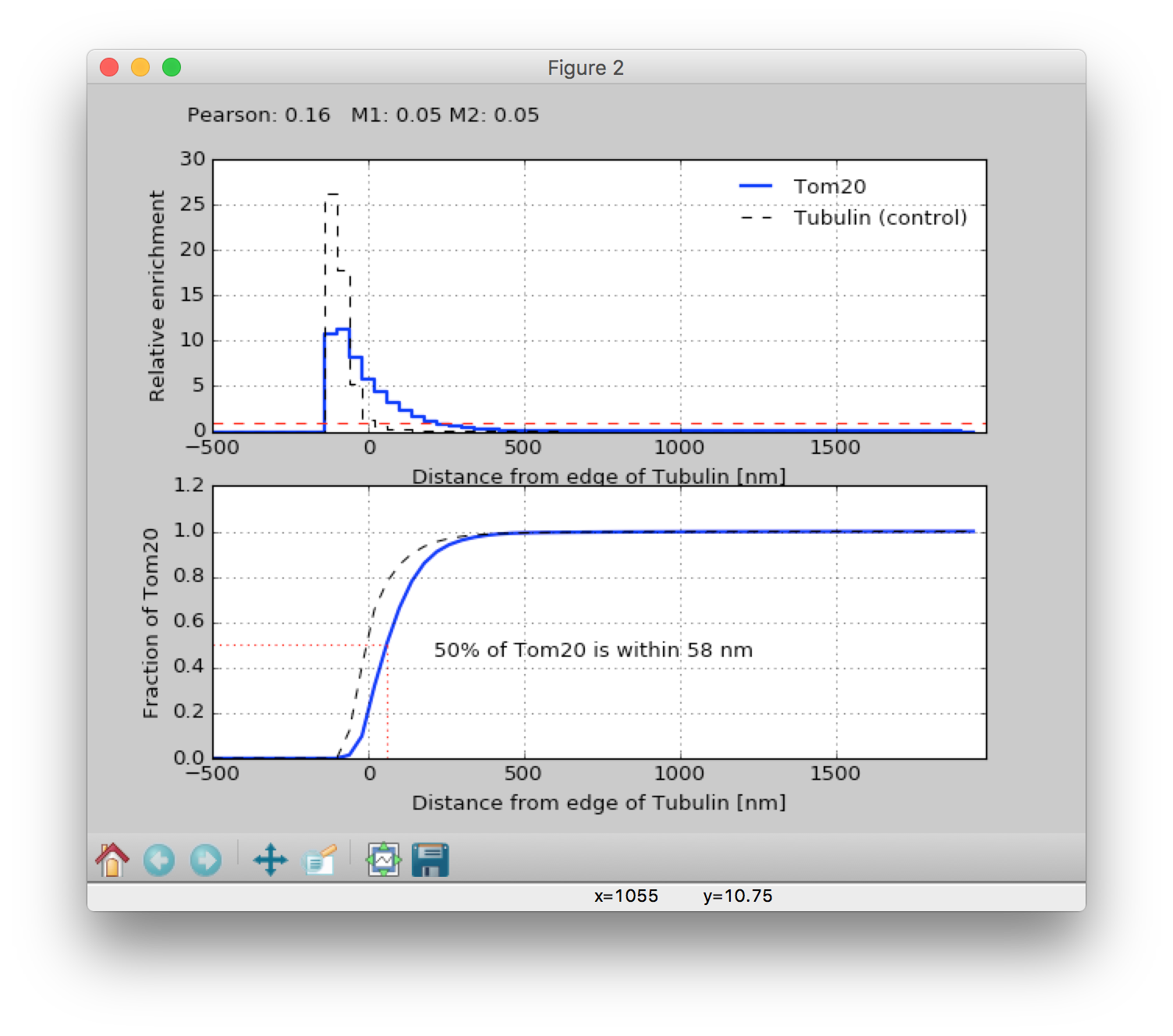
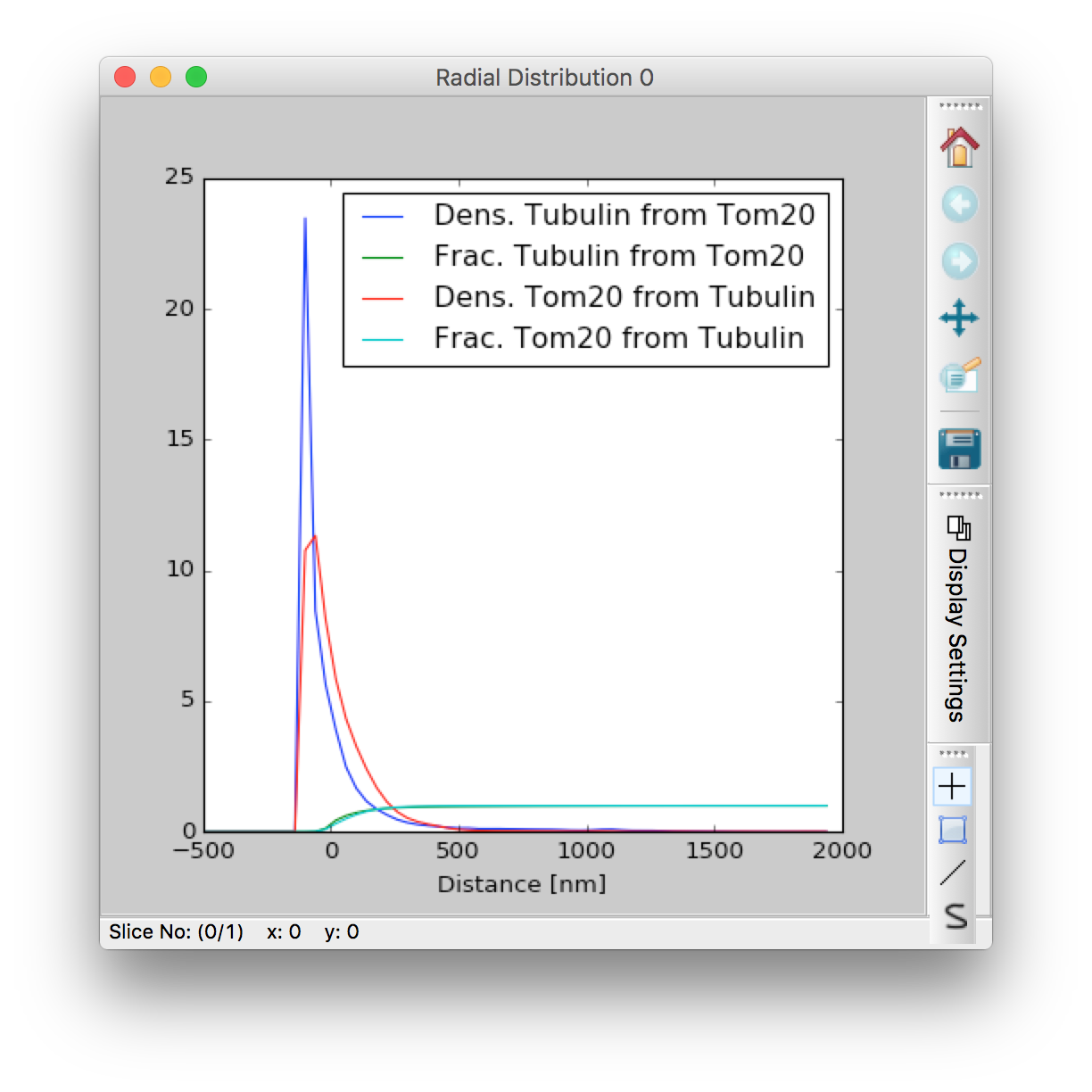
The first window shows the relative enrichment (comparing the density to an assumption of uniform spatial randomness) of label B at a given distance from label As mask (top panel), and the total fraction enclosed at a given distance (bottom panel). Negative distances are inside the mask, and Manders and Pearson coefficients are displayed at the top of the figure. The 50% of X within Ynm metric is my candidate for a new colocalization metric which will still work for super-resolution methods where nothing really colocalizes. The dotted line shows a comparison of same analysis performed on the raw intensities in the channel used to generate the mask, and essentially functions as a control for how good the thresholding is. The second window is a repeat of the first with the labels switched, and the 3rd window just displays the raw histogram data. The 3rd window is mainly interesting if you want to access the raw histogram data, which can be saved in a format which can be imported into Excel by bring this window to the front and then selecting File-> Save as from the menu. NOTE: when saving histograms you must set the `File type` in the Save as dialog to `Tab formatted text - .txt`.